Sara SATTIN
CV
Sara graduated in 2006 from the Università degli Studi di Milano in Industrial Chemistry and Management with full marks with honours (110/110 e lode). She obtained her PhD in Chemical Sciences in 2009 working on the synthesis of inhibitors of DC-SIGN mediated infections under the supervision of prof. Anna Bernardi.
In 2010 she joined the group of Prof. Javier de Mendoza at the ICIQ (Institute of Chemical Research of Catalonia, Tarragona, Spain) as a postdoctoral fellow working on voltage gated Potassium channel selective ligands and UOF (Uranyl Organic Frameworks) supramolecular cages (see Nat. Commun. 2012, 3, 785).
In 2012 she moved to Oxford (UK) to join the group of Prof. Ben G. Davis as a postdoctoral fellow in chemical biology, where she achieved the functional deconvolution of a highly diverse sugar library derived from the Formose reaction and worked towards the development of a Turing Test for protocells.
At the end of 2013 she returned to Milan where she is currently working on the design and synthesis of small molecular probes for the allosteric modulation of the chaperone protein Hsp90.
Since April 2017 she is Assistant Professor (RTD A) at UNIMI.
Research Interests
I am interested mainly in applying organic chemistry to chemical biology problems and molecular recognition events. This translates in the design and synthesis of:
- Glycomimetics, both at the mono- and multivalent level, aimed to interact with human lectins (e.g. DC-SIGN and MBL, see for example ACS Chem. Biol. 2010, 5, 301; Bioconjugate Chem. 2011, 22, 1354; Chem. Eur. J. 2016, 22, 3686 and Trends Biotechnol. 2016, 34, 483)
- Allosteric modulators of the chaperone protein Hsp90
Hsp90 (heat shock protein 90) is a chaperone protein that assists the correct folding of other proteins, called clients. Hsp90 plays a pivotal role in the cell life cycle and it is crucial for the cell response to stressful conditions. As many Hsp90 clients are oncoproteins, this chaperone is nowadays an established target in cancer therapy. From a structural point of view, Hsp90 is a homodimer and each monomer is constituted by three domains, an N-terminal Domain (NTD), where the main ATP-binding site is located, a Middle Domain (MD) and a C-terminal Domain (CTD), where the dimerization takes place.
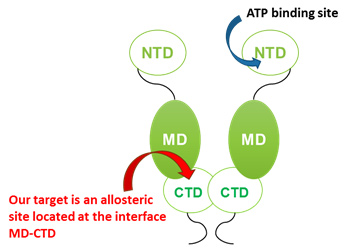
Schematic representation of the Hsp90 dimer and its structural domains
The majority of the small molecules developed targeting Hsp90 aim to displace the ATP from its binding site, blocking its ATPase and therefore chaperoning activity altogether. Unfortunately, this block triggers the Heat shock cell-survival mechanism, with increased expression of Heat shock proteins, including Hsp90 and Hsp70.
Our aim is instead to tune the activity of the chaperone with molecules targeting an allosteric site located between the MD and the CTD, close to the dimerization interface and recently discovered by Morra et al (J. Chem. Theory Comput. 2010, 6, 2978–2989).
We synthesised a family of diversified compounds based on the natural product Eupomatenoid-2 (Carbohydr. Res. 2014, 390C, 33-41) that are able to accelerate the ATPase activity of Hsp90 (Chem. Eur. J. 2015, 21, 13598-13608) with downstream effects that we are just beginning to unravel.
FRET assays and MD simulations support our hypothesis that the ATPase acceleration is due to a faster dimer closure rate and to an increased population of the catalytically active, so-called, closed-twisted conformation (Sci. Rep. 2016, 6, 23830).
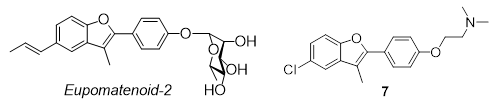
The lead compound Eupomatenoid-2 and one of our compounds, 7.
Data from STD-NMR studies carried out on 7 in the presence of Hsp90 validate our hypothesised binding mode, where the dimethyl-amino group of 7 is in proximity of two negatively charged residues (Asp503 and Glu477) (Eur. J. Org. Chem. 2016, 2016, 3349-3364).
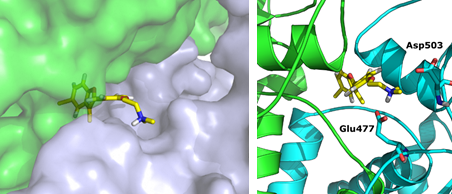
Hypothesised bindig model of compound 7 at the allosteric site
Publications
| Orcid ID |
* Corresponding author
24. The small RNA ReaL: a novel regulatory element embedded in the Pseudomonas aeruginosa quorum sensing networks S. Carloni; R. Macchi; S. Sattin; S. Ferrara; G. Bertoni
Environ Microbiol 2017, 19, 4220
23. Chaperones rescue the energetic landscape of mutant CFTR at single molecule and in cell
M. Bagdany; G. Veit; R. Fukuda; R. G. Avramescu; T. Okiyoneda; I. Baaklini; J. Singh; G. Sovak; H. Xu; P. M. Apaja; S. Sattin; L. K. Beitel; A. Roldan; G. Colombo; W. Balch; J. C. Young; G. L. Lukacs
Nature communications 2017, 8, 398
22. Design of Allosteric Stimulators of the Hsp90 ATPase as New Anticancer Leads I. D'Annessa, S. Sattin, J. Tao, M. Pennati, C. Sànchez-Martìn, E. Moroni, A. Rasola, N. Zaffaroni, D.A. Agard, A. Bernardi, and G. Colombo Chem. Eur. J. 2017, 23, 5188
21. Stereoselective innovative synthesis and biological evaluation of new real carba analogues of minimal epitope Manα(1,2)Man as DC-SIGN inhibitors V. Bordoni; V. Porkolab; S. Sattin; M. Thépaut; I. Frau; L. Favero; P. Crotti; A. Bernardi; F. Fieschi; V. Di Bussolo
RSC Adv. 2016, 6, 89578
20. Linear biocompatible glyco-polyamidoamines as dual action mode virus infection inhibitors with potential as broad-spectrum microbicides for sexually transmitted diseases N. Mauro; P. Ferruti; E. Ranucci; A. Manfredi; A. Berzi; M. Clerici; V. Cagno; D. Lembo; A. Palmioli; S. Sattin
Sci. Rep. 2016, 6, 33393
19. Synthesis of functionalized 2-(4-hydroxyphenyl)-3-methylbenzofuran allosteric modulators of Hsp90 activity S. Sattin;* M. Panza; F. Vasile; F. Berni; G. Goti; J. Tao; D. Agard; G. Colombo; A. Bernardi
Eur J Org Chem 2016, 2016, 3349
18. Molecular Dynamics Simulations Reveal the Mechanisms of Allosteric Activation of Hsp90 by Designed Ligands G. Vettoretti; E. Moroni; S. Sattin; J. Tao; D. A. Agard; A. Bernardi; G. Colombo
Sci Rep 2016, 6, 23830
17. Glycoconjugates and Glycomimetics as Microbial Anti-Adhesives S. Sattin; A. Bernardi
Trends Biotechnol. 2016, 34, 483
16. Design and synthesis of glycomimetics S. Sattin; A. Bernardi In Carbohydrate Chemistry: Volume 41; The Royal Society of Chemistry: 2016; Vol. 41, p 1
15. Detection and quantitative analysis of two independent binding modes of a small ligand responsible for DC-SIGN clustering C. Guzzi; P. Alfarano; I. Sutkeviciute; S. Sattin; R. Ribeiro-Viana; F. Fieschi; A. Bernardi; J. Weiser; J. Rojo; J. Angulo; P. M. Nieto
Org Biomol Chem 2016, 14, 335
14. Scaffold Optimisation of Tetravalent Antagonists of the Mannose Binding Lectin G. Goti; A. Palmioli; M. Stravalaci; S. Sattin; M. G. De Simoni; M. Gobbi; A. Bernardi
Chem. Eur. J. 2016, 22, 3686
13. Activation of Hsp90 Enzymatic Activity and Conformational Dynamics through Rationally Designed Allosteric Ligands S. Sattin; J. Tao; G. Vettoretti; E. Moroni; M. Pennati; A. Lopergolo; L. Morelli; A. Bugatti; A. Zuehlke; M. Moses; T. Prince; T. Kijima; K. Beebe; M. Rusnati; L. Neckers; N. Zaffaroni; D. A. Agard; A. Bernardi; G. Colombo
Chem. Eur. J. 2015, 21, 13598 highlighted as Very Important Paper
12. DC-SIGN as a Target for Drug Development Based on Carbohydrates
S. Sattin; F. Fieschi; A. Bernardi In Carbohydrate Chemistry: State of the Art and Challenges for Drug Development; IMPERIAL COLLEGE PRESS: 2015, p 379
11. Carbohydrates: A phenol sandwich fights diabetes A. Bernardi; S. Sattin
Nat Chem Biol 2015, 11, 635
10. Unique DC-SIGN clustering activity of a small glycomimetic: A lesson for ligand design I. Sutkeviciute; M. Thepaut; S. Sattin; A. Berzi; J. McGeagh; S. Grudinin; J. Weiser; A. Le Roy; J. J. Reina; J. Rojo; M. Clerici; A. Bernardi; C. Ebel; F. Fieschi
ACS Chem Biol 2014, 9, 1377
9. Synthesis of potential allosteric modulators of Hsp90 by chemical glycosylation of Eupomatenoid-6 L. Morelli; A. Bernardi; S. Sattin*
Carbohydr Res 2014, 390C, 33
8. Pseudo-Mannosylated DC-SIGN Ligands as Potential Adjuvants for HIV Vaccines A. Berzi; N. Varga; S. Sattin; P. Antonazzo; M. Biasin; I. Cetin; D. Trabattoni; A. Bernardi; M. Clerici
Viruses 2014, 6, 391
7. Structure of a glycomimetic ligand in the carbohydrate recognition domain of C-type lectin DC-SIGN. Structural requirements for selectivity and ligand design M. Thepaut; C. Guzzi; I. Sutkeviciute; S. Sattin; R. Ribeiro-Viana; N. Varga; E. Chabrol; J. Rojo; A. Bernardi; J. Angulo; P. M. Nieto; F. Fieschi
J Am Chem Soc 2013, 135, 2518
6. Giant regular polyhedra from calixarene carboxylates and uranyl S. Sattin; S. Pasquale; E. C. Escudero-Adan; M. Martinez-Belmonte; J. de Mendoza
Nat. Commun. 2012, 3, 785 highlighted in ChemistryViews
5. Design, synthesis and activity evaluation of mannose-based DC-SIGN antagonists N. Obermajer; S. Sattin; C. Colombo; M. Bruno; U. Švajger; M. Anderluh; A. Bernardi
Mol Divers 2011, 15, 347
4. Pseudosaccharide functionalized dendrimers as potent inhibitors of DC-SIGN dependent Ebola pseudotyped viral infection J. Luczkowiak; S. Sattin; I. Sutkeviciute; J. J. Reina; M. Sanchez-Navarro; M. Thepaut; L. Martinez-Prats; A. Daghetti; F. Fieschi; R. Delgado; A. Bernardi; J. Rojo
Bioconjugate Chem. 2011, 22, 1354
3. Inhibition of DC-SIGN-mediated HIV infection by a linear trimannoside mimic in a tetravalent presentation S. Sattin; A. Daghetti; M. Thepaut; A. Berzi; M. Sanchez-Navarro; G. Tabarani; J. Rojo; F. Fieschi; M. Clerici; A. Bernardi
ACS Chem. Biol. 2010, 5, 301
2. An assay for functional dendritic cell-specific ICAM-3-grabbing nonintegrin (DC-SIGN) inhibitors of human dendritic cell adhesion N. Obermajer; U. Svajger; M. Jeras; S. Sattin; A. Bernardi; M. Anderluh
Anal. Biochem. 2010, 406, 222
1. 1,2-Mannobioside mimic: synthesis, DC-SIGN interaction by NMR and docking, and antiviral activity J. J. Reina; S. Sattin; D. Invernizzi; S. Mari; L. Martinez-Prats; G. Tabarani; F. Fieschi; R. Delgado; P. M. Nieto; J. Rojo; A. Bernardi
ChemMedChem 2007, 2, 1030
Patent
Inhibitors of microbial infections (2011). A. Bernardi, S. Sattin, M.S. Clerici, AMV Berzi, F.J. Rojo, M. Sanchez, J.J. Reina, F. Fieschi Inhibitors of microbial infections PCT Int. Appl., WO 2011000721 A1 20110106

