Martino Bolognesi has been one of the founders of X-ray structural biology in Italy where, over the years, he opened labs at the Universities of Pavia, Genova and Milano. The research groups that are linked in this web site, and operate as independent units sharing local infrastructures and resources, originated since his transfer at the University of Milano in 2005. Besides his senior supervisor role, Prof. Bolognesi maintains an active interest in the application of X-ray structural biology to Structural Vaccinology and to the definition of open structural-functional questions in heme-proteins and enzymes. In his career he has served in several scientific advisory boards and synchrotron facilities in Italy and in Europe (ESRF, EMBL-Heidelberg, EMBL-Hamburg, ESRF, ELETTRA...). Martino Bolognesi is EMBO member, and member of Academia Europea, Istituto Lombardo di Scienze e Lettere, Accademia Nazionale dei Lincei.
Since 2016 he has been a leading figure in the set-up of the new single particle cryo EM facility that is run at the University of Milano in collaboration with the Pediatric Clinical Research Center (Fondazione Romeo e Enrica Invernizzi - Milano).

Selected Publications
Cryo-EM structure of cardiac amyloid fibrils from an immunoglobulin light chain AL amyloidosis patient. Swuec P, et al. Nat Commun. 2019 (PubMed: 30894521)
Cas9 Allosteric Inhibition by the Anti-CRISPR Protein AcrIIA6. Fuchsbauer O, Swuec P, et al. Mol Cell. 2019 (PubMed: 31604602)
Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Nardini M, et al. Cell. 2013 (PubMed: 23332751)
Structure and functionality in flavivirus NS-proteins: perspectives for drug design. Bollati M, et al. Antiviral Res. 2010. (PubMed: 19945487)
Current Group

Scientific Programmes
Cryo-EM
READy
EGV
The Cryo-EM Laboratory focusses on the architectural characterization of macromolecular complexes as targets for a deeper understanding of the molecular bases of disease; as a result of such structural investigations, the mechanisms of drug action can be dissected, and the design of innovative drugs carried over. Pivotal to the experimental investigations carried out in the lab is the installation of a new FEI Talos Arctica 200 kV FEG electron microscope equipped with a state-of-the-art FEI Falcon 3EC direct electron detector and Volta Phase-plate. The Cryo-EM Lab also benefits from a dedicated sample preparation room to ensure the highest specimen quality and a GPU-based computing cluster for efficient and hustle-free data analysis. The Cryo-EM Lab aims at unraveling the three-dimensional structure and dynamics of large molecular machines at sub-nanometer resolution. The installation, the first of this kind in Italy, is supported by the University of Milano and by Fondazione Invernizzi. Current lab staff are Prof. Martino Bolognesi and Dr. Paolo Swuec. A technician position is scheduled for the end of 2017.

A REgionAl network for the development of rapid Diagnostic tools to address emerging epidemics and bio-emergencies. (READy)
The aim of READy is to form a regional, scientific network of excellence for the rapid response to bio-emergencies. The project focuses on the development of new bioreagents and diagnostic kits that target emerging tropical diseases, including Chagas disease, diseases caused by arboviruses, Malaria and Schistosomiasis.
Epitope peptides for immunodiagnostic use will be predicted and designed using computational methods, starting from the crystal structures of known seroreactive Schistosoma and Trypanosoma antigens. UNIMI is actively producing recombinant Schistosoma and Trypanosoma antigens for immunological tests and 3D structure characterisation.
Funding:
Regione Lombardia (2017-2020) PI-Prof Bolognesi
Collaborators:
ICRM-CNR, Diagnostic Probes Srl., PRIMM Srl., Universita di Bicocca, Politecnico di Milano, Istituto Nazionale di Genetica Molecolare, Ospedale Luigi Sacco di Milano.

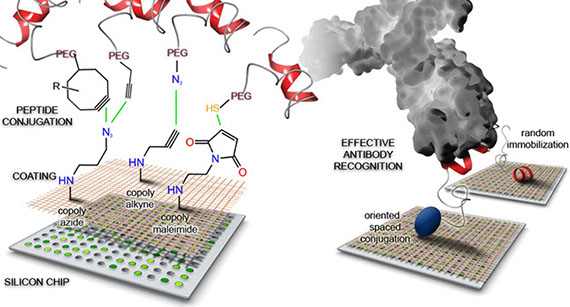
Epitope peptide-based microarray platforms for rapid immunodiagnostics. Epitope peptides are predicted from 3D antigen structures and subsequently synthesised as free peptides. Peptides are immobilised in specific orientations on polymeric coated microchips using click chemistry. This figure is from Gori et al., 2016 (DOI: 10.1021/acs.bioconjchem.6b00426)
Epitope grafting on bacterial vesicles to develop a novel Burkholderia Vaccine.
Research efforts regard X-ray crystallography studies of protein antigens as targets for computational epitope mapping and vaccine component design. We target antigens from the Gram-negative bacterium Burkholderia pseudomallei that causes melioidosis, responsible for significant mortality in endemic populations. Using epitope grafting, we aim to generate 'super antigens' that host multiple epitopes, in order to induce an enhanced immune response. To validate our efforts, we will display the generated epitopes, Ags and grafts on Outer Membrane Vesicles, which will be exploited as multi-epitope presentation vessels for in vivo tests.
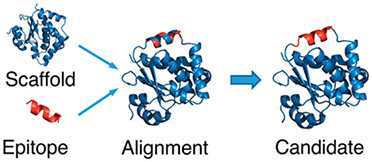
llustrazione of epitope grafting. An epitope from one antigen is transplanted onto a structurally apt region of another antigen or heterogenous protein scaffold. This figure is from Capelli et al., 2017 (DOI: 10.1021/acs.jcim.6b00584)
Funding: PRIN Contract number 2015JTL4HL (2016-2019). PI-Prof Bolognesi
Collaborators: Dr. G. Colombo (ICRM-CNR, Milano), Dr. G. Grandi (University of Trento).
The Structural Biology Group comprises members from both the DBS-UNIMI and the IBF-CNR. The content herein is not regulated by the University of Milan.



