Regulatory networks at the interface between environmental cues and reproductive development
 Rice
(Oryza sativa) is an important model system for many plant scientists
and has been instrumental to many key discoveries in biology. Similarly to many
plant species, the onset of its reproductive phase takes place during a
specific time of the year, when environmental conditions are optimal for inflorescence
development, fertilization and seed formation. The switch to reproductive
growth depends upon measurement of suitable external parameters, among which
day length (also called photoperiod) represents the most predictable one.
Rice
(Oryza sativa) is an important model system for many plant scientists
and has been instrumental to many key discoveries in biology. Similarly to many
plant species, the onset of its reproductive phase takes place during a
specific time of the year, when environmental conditions are optimal for inflorescence
development, fertilization and seed formation. The switch to reproductive
growth depends upon measurement of suitable external parameters, among which
day length (also called photoperiod) represents the most predictable one.
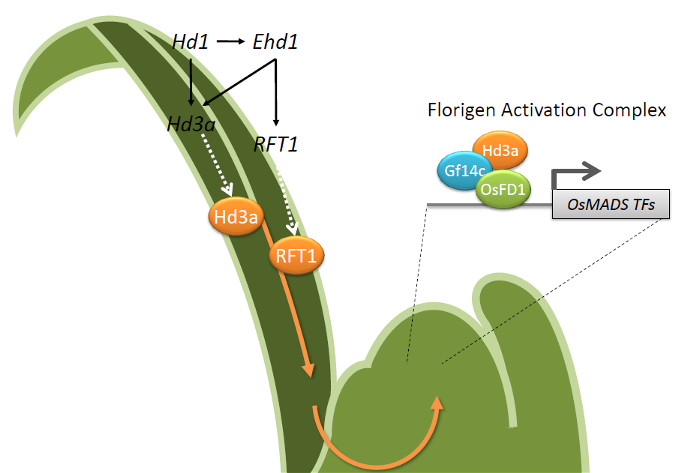
Flowering of rice is promoted upon exposure to short days, when the length of the night exceeds a critical threshold. These conditions promote transcription of HEADING DATE 3a (Hd3a) and that of its paralog, RICE FLOWERING LOCUS T 1 (RFT1) in the phloem of leaves. Both genes encode for “florigens”, small, systemic protein signals that move through the vascular stream to reach the shoot apical meristem. At the apex, they determine its developmental conversion into an inflorescence.
Gene networks operating in leaves are necessary to define spatial and temporal expression patterns of Hd3a and RFT1. To this end, several regulatory proteins, including transcription factors, protein kinases and F-box proteins transduce environmental information into florigenic molecular signals. Prominent among such regulators are HEADING DATE 1 (Hd1) and EARLY HEADING DATE 1 (Ehd1), both of which encode transcription factors promoting expression of the florigens under inductive short days.
Downstream of Hd3a and RFT1, molecular networks at the shoot apical meristem perceive the arrival of the florigens and initiate a series of molecular events that lead to activation of inflorescence identity genes. Central in this mechanism is a heterohexameric complex called Florigen Activation Complex (FAC), whose DNA binding at target loci is determined by transcription factors of the bZIP family. Assembly and activation of the FAC ultimately results in expression of OsMADS transcription factors that drive inflorescence formation.
Our lab aims at isolating novel components of gene regulatory networks controlling expression of the florigens in leaves, as well as response of the shoot apical meristem to florigenic signals. We defined a key regulatory step in leaf networks, by showing that Hd1 acts as repressor of Ehd1 in leaves (Gomez-Ariza et al., 2015). This node ensures that under non-inductive long days, rice flowering is prevented or strongly delayed. We explored the effects of drought stress on flowering, showing that it causes a delay in the onset of the reproductive phase, by interacting with components of the leaf photoperiodic network (Galbiati et al., 2016). Finally, we first identified and functionally described florigen-containing complexes that limit Ehd1 expression, to self-tune florigen production (Brambilla et al, 2017).
Rice adaptation to higher latitudes and breeding
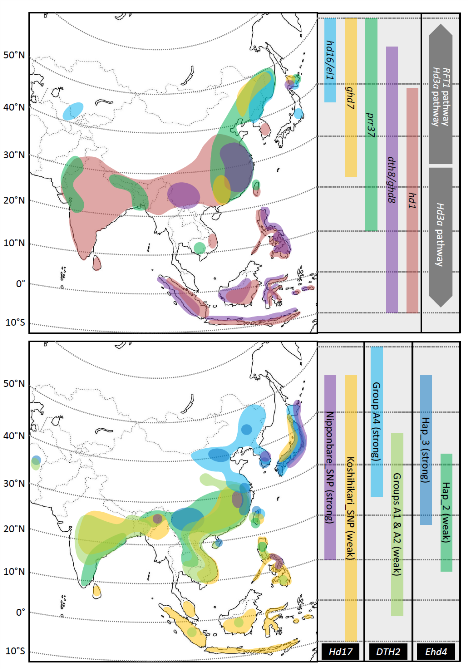
Rice was domesticated in Southern China about 10.000 years ago. Its cultivation subsequently expanded in all continents, and currently rice can be grown across a large latitudinal range, between 36°S and 55°N. During the fifteenth century rice reached Europe and its cultivation became established around the Mediterranean Area. Currently, Italy is the major European rice producer.
Because of its tropical origin, cultivation of rice at northern latitudes required extensive genetic adaptation. Sensitivity to the photoperiod has been a major obstacle to expansion, because rice flowers most effectively when grown under short days that are not typical of Mediterranean summers. Artificial selection has increased the frequency of specific alleles (usually loss-of-function alleles of floral-repressor genes), that accelerate flowering when rice is grown under non-inductive long days. The Figure on the left (from Shrestha et al., 2014) represents a possible distribution of such alleles in Asia.
Our
lab is interested in understanding how modifications of the photoperiodic
network allowed rice to flower also under long days. Using rice varieties
cultivated in Europe, we explored the extent of natural variation at loci
controlling sensitivity to the photoperiod (Gomez-Ariza et al., 2015). We
uncovered an unexpected high rate of mutations at photoperiodic flowering
genes, as well as extensive pyramiding of loss-of-function alleles (Goretti et
al., 2017). Such variation largely explains the flowering behavior of rice
accessions cultivated in Europe, whose cycle length perfectly matches the length
of the cropping season. It further provides the necessary conceptual framework
to drive selection of novel genotypes.