Selected Publications
Planchestainer M, Hegarty E, Heckmann CM, Gourlay LJ, Paradisi F. Widely applicable background depletion step enables transaminase evolution through solid-phase screening. Chem Sci. 2019 May 9;10(23):5952-5958. doi: 10.1039/c8sc05712e. PMID: 31360401
De Vitis V, Nakhnoukh C, Pinto A, Contente ML, Barbiroli A, Milani M, Bolognesi M, Molinari F, Gourlay LJ, Romano D. A stereospecific carboxyl esterase from Bacillus coagulans hosting nonlipase activity within a lipase-like fold. FEBS J. 2018 Mar;285(5):903-914. doi: 10.1111/febs.14368. Epub 2018 Jan 11. PMID: 29278448
Structure and Computation in Immunoreagent Design: From Diagnostics to Vaccines. Gourlay L, Peri C, Bolognesi M, Colombo G. Trends Biotechnol. 2017. [PubMed: 28739221]
Champion, OL et al. (2016) Immunisation with proteins expressed during chronic murine melioidosis provides enhanced protection against disease. Vaccine. 34(14):1665-71. [PubMed: 26917010]
Gourlay, L.J. et al. (2015) From crystal structure to in silico epitope discovery in Burkholderia pseudomallei flagellar hook-associated protein FlgK. FEBS J. 282(7), 1319-33. [PubMed: 25645451]
Current Group
Rachele Sala
MS Student

Scientific Programmes
SMART
NFIX
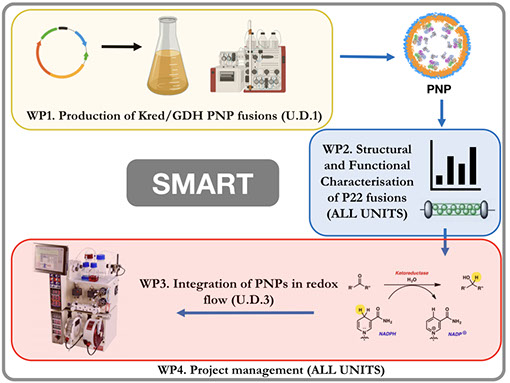
The SMART project will develop an innovative and robust platform for the sustainable, high-yield, lab-scale preparation of active pharmaceutical ingredients or their key intermediates. Specially, SMART will generate a protein-based nanoparticle (PNP) system that encapsulates a ketoreductase (KRED) within the Salmonella bacteriophage P22 capsid, together with glucose dehydrogenase (GDH) for continuous, cost-effective NADPH recycling.
SMART will exploit state-of-the art molecular biology, biochemistry, structural biology and chemistry methods to encapsulate KRED and GDH within the Salmonella bacteriophage P22 capsid. 300 copies of the P22 scaffolding protein (SP) self-assemble with 420 copies of a coat protein, to form the P22 nanocage, thus fusion of our enzyme couple to SP will result in equally high copy numbers enclosed within the P22 capsid interior. PNP fusions will be structurally and functionally characterised and used in an optimised chemical flow setup. As proof-of-principle, we will validate our innovative PNP redox flow system with the synthesis of a series of enantiomerically pure, pharmaceutically relevant alcohols.
Self-assembling Multi-enzyme nAnoparticles for continuous Redox biocaTalysis (SMART)
Funding:
SEED 4 Innovation, (Linea 3 Università degli Studi di Milano)
Collaborators:
Project in collaboration with Prof. D. Romano (DEFENS) e Dr. L. Tamborini (DISFARM).
"Production of full-length Nuclear Factor 1 X in the Leishmania tarantolae expression system (LEXSY)"
I have been awarded a departmental grant to produce the full-length form of the transcription factor Nfix (Nuclear factor 1 X) in the Leishmania tarantolae eucaryotic expression system. Nfix is of clinical interest due to its role in a number of human cancers and disease-specific mutations in the nfix gene give rise to two debilitating, congenital malformation syndromes: Marshall-Smith Syndrome (MSS) and Sotos-like Syndrome (Malan Syndrome). Both pathologies share similar clinical features including, accelerated skeletal anomalies, physical malformations and intellectual retardation. Interestingly, disease-specific mutations of both syndromes are clustered into distinct regions of the nfix gene.
The availability of a full-length Nfix construct will allow a plethora of structural and functional studies to be carried out, in order to gain insight into the molecular bases of Nfix-DNA recognition. In light of the known location of disease-specific mutations responsible for Malan Syndrome and MSS, production of full-length Nfix will allow us to study the structure-function consequences on DNA binding of the disease specific mutations that lead to these syndromes.
Funding: Universita' degli Studi di Milano 'Linea 2' funding 2020-2021
The Structural Biology Group comprises members from both the DBS-UNIMI and the IBF-CNR. The content herein is not regulated by the University of Milan.




